SpatialMeta (Online MALDI data analysis server)
A online interactive website for analyzing MALDI and spatial metabolomics/proteomics data.
Introduction
MALDI MSI, also known as matrix-assisted laser desorption/ionization mass spectrometry imaging, is a an instrument to evaluate the spatial distribution of analytes (proteins, peptides, medicines, lipids, and metabolites) in thin tissue sections.
Step 1
- Data format convertion
We have tested datas from the Bruker MALDI-TOF system. Data from Bruker MALDI-TOF system can be easily exported as .imzML and .ibd format. Our SpatialMeta accept these two input formats for downstream analysis pipeline.
You may need imzMLConverter, in order to get ready, you need to do as following:
- Download the latest version of imzMLConverter
- Install Java SE 8 JRE
- Install ProteoWizard
- [Optional] Install MS Data Converter (SCIEX) to enable conversion of data from AB SCIEX data instruments
Step 2
- Data upload
Step 3
- Start your analysis
Vendor instruments
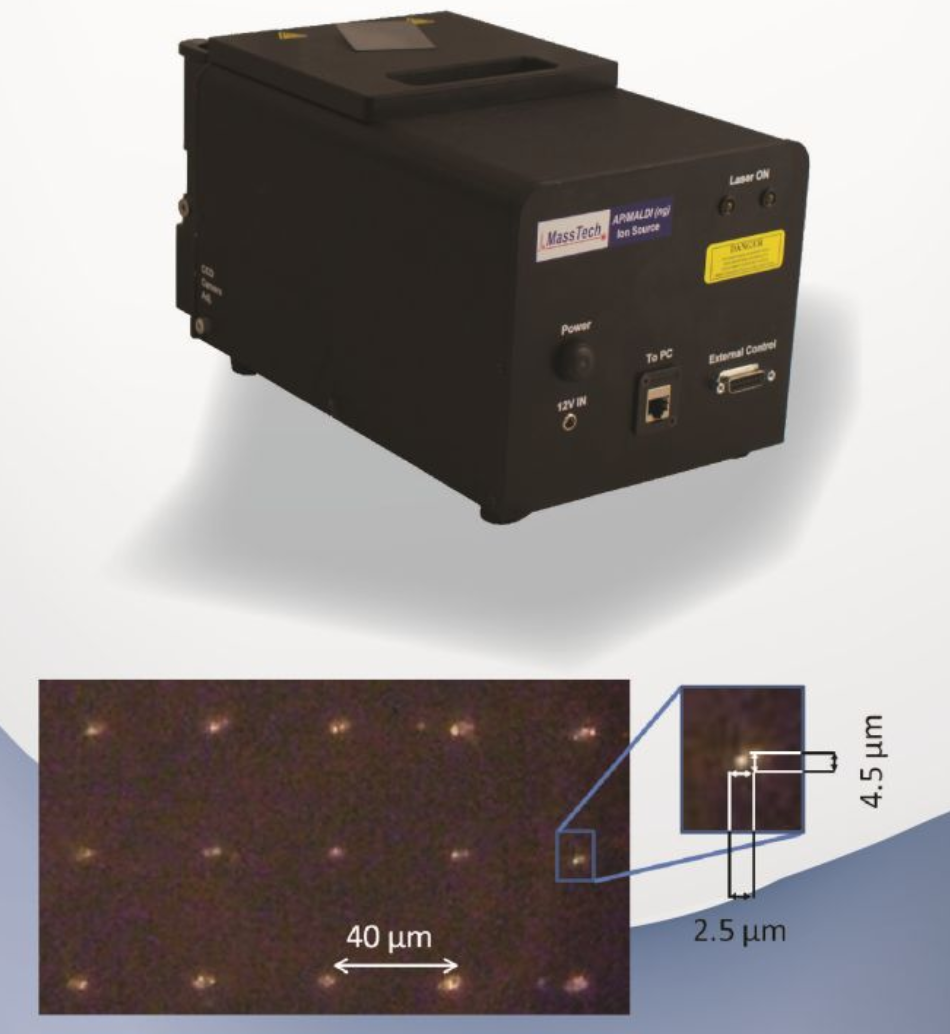
- AP/MALDI (ng) UHR, MassTech
- The AP-MALDI(ng) UHR is available for all mass spectrometers currently supported by the regular AP-MALDI. The AP-MALDI(ng) UHR includes upgraded optics, source housing and eliminates the use of optical fiber which allow for a focused laser spot less than 10 micron in diameter.
- AP-MALDI(ng) UHR includes an upgraded laser – it is still a solid state Nd:YAG laser at 355 nm wavelength, but the output frequency is up to 10 kHz.
- rapifleX®, MALDI-TOF/TOF system, Bruker
- Speed, Make the best decisions in the shortest amount of time with the 10 kHz scanning smartbeam 3D laser, for throughput up to 20 times faster than traditional MALDI-TOF systems.
- Robustness, Most robust, yet easy to access ion source enables straightforward cleaning, designed for maximum uptime in 24/7 operations.
