Tailoring the brain connectivity atlas: reconstruction and revision of large-scale microscopic brain atlas
In complex biological systems, it is often the structure of the object of study that determines its function. This is also true for neural network systems. In order to understand the complexity of the structure of animal brain networks and their potential functions, brain mapping is one of the important goals in the current brain project.
On August 19, 2019, Beijing Center for Brain Science and Brain-like Research invited Dr. Ting Zhao from Janelia Institute to give an exciting lecture on “Brain Mapping Reconstruction and Correction”.
 Figure 1: Dr. Ting Zhao in the lecture
Figure 1: Dr. Ting Zhao in the lecture
Compared to sparse reconstruction based on optical imaging (for the region of interest), dense reconstruction based on electron microscopy (reconstructing all distinguishable neurons within the sample) is extremely laborious, and Janelia FlyEM’s team recognized from the beginning the importance of dense reconstruction, which is the only way to fully reproduce the complexity of the nervous system.
Dr. Ting Zhao from Janelia FlyEM team firstly introduced the whole FlyEM team in his presentation. FlyEM project team is currently the largest project team in Janelia with about 70 members, including Imaging scientists, Computational Experts, Biologists, The goal of the FlyEM team is to reconstruct the entire Drosophila nervous system. The task of reconstructing the brain atlas of synaptic-level structures from such dense electron microscopic images, for which raw data are currently obtained by electron microscopy, is unusual in terms of the amount of work involved and the challenges it poses because of the different disciplines involved.
The Janelia FlyEM project team has been working on this project for 10 years, from the first column of the Drosophila cerebellum (379 neurons) [1] to seven columns (~1000 neurons) [2], to the neuronal remodeling of the Drosophila mushroom body (~1000 neurons) [3]. The completion time of the project was gradually reduced from 5 years to 2 years, and finally all the neurons in the Mushroom body were reconstructed in only 1 year. With the breakthroughs and innovations of the team, the overall efficiency of the FlyEM team has been rapidly improved to support larger scale brain atlas reconstruction work. Recently, FlyEM continues to challenge new heights, aiming at the construction of half of the Drosophila brain atlas.
The whole process of brain atlas reconstruction is as follows: sample preparation → electron microscopy imaging → image processing → image correction, and finally morphological and connectomic analysis.
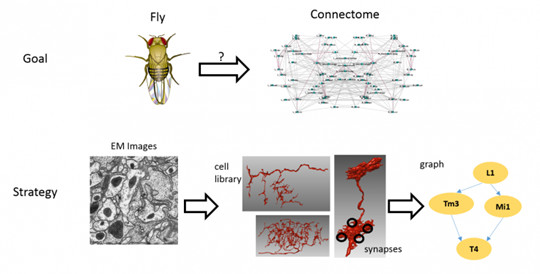 Figure 2: Janelia FlyEM team project - Drosophila dense brain mapping reconstruction[4]
Figure 2: Janelia FlyEM team project - Drosophila dense brain mapping reconstruction[4]
At present, image segmentation is an important task in brain mapping reconstruction. Dr. Ting Zhao mentioned that the FlyEM team’s image segmentation work at a later stage is in collaboration with Google team, using their latest algorithm FFN (Flood-Filling Networks) to automatically perform image segmentation and reconstruction of neurons on Drosophila electron microscopy imaging data to rapidly build brain atlas. [5]
Dr. Ting Zhao is the main person in charge of the “brain atlas correction process”. At this point, the interesting question is, how far is the automatic processing result from the real brain atlas? Dr. Ting Zhao describes it as “a dead horse on a hill”. As you can see, the distance is still very far, so how can we reach the real brain atlas quickly? At this point, an efficient correction software is extremely important. The main work of Ting Zhao’s team is to solve this major problem in the whole brain atlas construction process, i.e. to develop software to speed up the manual correction of the automatic processing results and to improve the efficiency of the brain atlas drawing work.
The development of correction software from the earliest generation of Raveler, to NeuTu, and then to Neu3 has experienced a qualitative leap in terms of data size, complexity of connection groups, and image segmentation quality. NeuTu and Neu3 provide a visual interface directly to the corrector, allowing the corrector to quickly locate errors, and DVID is an important part of the software, which is simply a distributed image database with the ability to upload data to Google Cloud.
In general, an important feature of these software is the ability to visualize in 3D and “make the connectome visible”, like a GPS navigation system that allows multi-scale visualization for neuronal morphology. Fast conversion and localization between 2D images and 3D visualizations are supported, as well as annotation in 2D and 3D windows.
There are two other problems in segmentation: over-segmentation and over-merging. The over-segmented ones need to be merged, and the over-merged ones need to be separated. Ting Zhao designed a general framework for multi-scale interactive segmentation based on the seed point algorithm. The framework does not depend on the specific segmentation algorithm and can be easily replaced by the seed point-based segmentation algorithm. The more regions are prompted, the more accurate its results will be. The superpixel-based clipping (Cleaving) tool can correct most of the errors. A superpixel is a combination of pixels, each superpixel if a node and a network to indicate that they are connected together. With the integration of the voxel separation and the superpixel-based Cleaving tool, the two swords can be combined to efficiently correct any over-merging errors.
In addition, the FlyEM team has developed a Focused proofreading algorithm specifically designed to correct over-segmentation, analysis, evaluation, importance calculation, and ranking, thus further improving the overall efficiency of the correction tool.
At the end of the talk, Dr. Ting Zhao shared a video of the effect of a modified neuron and looked forward to the possibility that his project group will continue to challenge larger model animals in the future.
References
【1】Takemura, Shin-ya, et al. “A visual motion detection circuit suggested by Drosophila connectomics.” Nature 500.7461 (2013): 175.
【2】Takemura, Shin-ya, et al. “Synaptic circuits and their variations within different columns in the visual system of Drosophila.” Proceedings of the National Academy of Sciences 112.44 (2015): 13711-13716.
【3】Takemura, Shin-ya, et al. “The comprehensive connectome of a neural substrate for ‘ON’motion detection in Drosophila.” Elife 6 (2017): e24394.
【4】https://www.janelia.org/project-team/flyem
【5】Januszewski, Michał, et al. “High-precision automated reconstruction of neurons with flood-filling networks.” Nature methods 15.8 (2018): 605.
Writing | Jun-Liszt Li
Editor | Jun-Liszt Li
Audit | Wenzhi Sun

